02-09-2015
Catarina Coelho, Teresa Santos Silva and Maria João Romão from Macromolecular Crystallography Laboratory at Faculdade de Ciências e Tecnologia of Universidade NOVA de Lisboa (FCT-NOVA) crystallized and determined the AOX 3D structure, after a long and laborious process, and the results can be used to improve drug design studies in order to produce better drugs in a shorter period of time and with less failures, thus decreasing their overall cost. This research work was done in collaboration with researchers from the University of Potsdam, and the results are published today, 31st of August, in Nature Chemical Biology.
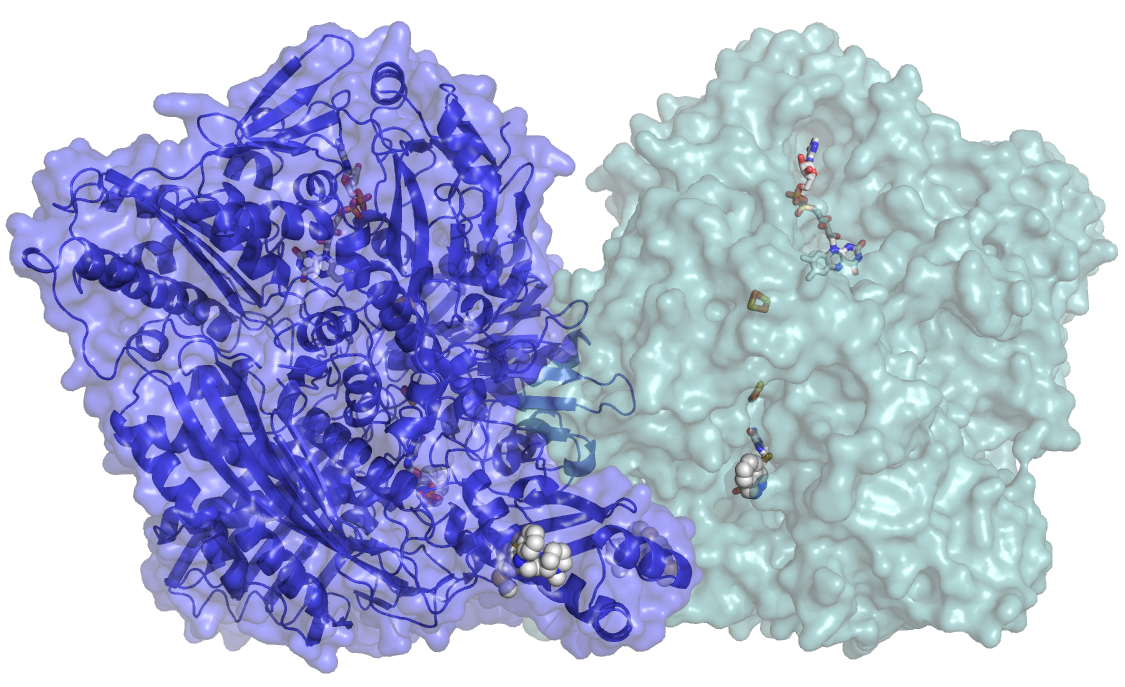
The enormous costs and efforts involved on drug research have a great impact in the pharmaceutical industry, especially due to the high rate of unsuccessful clinical research studies. One of the causes for the failure of clinical trials, where newly designed and synthesized drug molecules are being tested, is the unspecific degradation of the drug molecule by enzymes. Aldehyde Oxidase (AOX) is one of these enzymes, and its involvement in the metabolism of drugs has enormous impact in drug design justifying the great interest of the pharmaceutical industry in AOX.
AOX is present in the human liver and known to metabolize unspecific pharmaceutical drugs and xenobiotic compounds due to its broad range of substrates. As a direct consequence, drug design studies cannot predict if the new created drug molecules will be targets of the enzyme, and consequently degraded by AOX, leading to unsuccessful preclinical studies. Besides, the number of AOX enzymes differs in different animal species: while humans have only one, rodents are endowed with four, and therefore there is no suitable animal model to predict human AOX metabolism of new drug candidates before going into a clinical trial process.
“In this work we could crystallize and determine the 3D structure of AOX, providing important insight into catalytic and inhibition modes of the enzyme. This will enable to improve the drug design studies pipeline, allowing drug design companies to develop in silico models to predict AOX metabolism, which is essential to guide drug discovery and to screen for pharmacokinetics for clinical trials”, explains Maria João Romão, coordinator of the Macromolecular Crystallography research team and director of the UCIBIO research unit. The knowledge of the 3D structure of the enzyme will help to understand the existence of single point mutations (SNP) in the human enzyme, which are silent but have important consequences on the liver metabolism.
The complete protein has more than 1330 amino acid residues (more than 10 thousand atoms). “Due to its large dimension, our approach was to use X-Ray macromolecular crystallography to determine the 3D structure of the enzyme and to see the intrinsic and complicated network of amino acid arrangement. Only when the positions of most of the 10 thousand atoms were identified, could we correlate the structure with the function and draw conclusions regarding enzymatic and inhibition mechanisms”, explains Catarina Coelho, post-doc at FCT/UNL and first author of the article. Researchers of the Macromolecular Crystallography Laboratory work on AOX enzyme since eight years ago, but their studies were focused on the mouse enzyme that resulted in various publications, including its structure on The Journal of Biological Chemistry, in 2012. The human AOX contains complex metal cofactors (molybdenum and iron) that are not easily synthesized in bacteria, the host organism chosen to express enough quantity of the protein. A special system had to be developed, in collaboration with researchers in the University of Potsdam in Germany, so that the enzyme could be produced, in Escherichia coli, in sufficient amounts and with high degree of purity. In protein crystallography, “the bottleneck is to obtain suitable crystals; the experience we gained by working with the mouse AOX enzyme allowed us to start working on the human protein”, explains Maria João Romão.

In order to solve the AOX structure, more than 800 crystals had to be measured at several Synchrotron Radiation Labs: ESRF in France, DLS in the United Kingdom and SLS in Switzerland. During the past four years, “we needed more than 40 trips to the Synchrotron and many sleepless nights to get good data of AOX crystals”, says Catarina Coelho and Teresa Santos-Silva, FCT investigator (IF) and co-author of the article. With those results, the authors were able to solve the structure of the native enzyme and of a complex with an antipsychotic drug (thioridazine). “The data revealed a novel binding site that might be general for related drugs”, explains Teresa Santos-Silva, and “the major achievements described in our publication will allow us, in the near future, to develop new inhibitors as well as to design new drugs resistant to metabolism by AOX, helping in this way to reduce the failure of clinical trials”.

Mais notícias: